Saving these for later as I get into DeepCell–
I found this tool useful/essential for viewing TIFF files from wet lab data: QuPath
Objective?
DeepCell provides a pre-trained model, and tools to train models, for segmenting cells. Given a picture (e.g. from a microscope) identify which pixels are part of the same cell. The resulting information can be used for many sciency things that I don’t understand.
The library works great at small data sizes (512², 1024²), a few MB, but “real life” data is hundreds of MBs if not more.
The overall objective is to understand how DeepCell performs with huge images and how to make a pipeline to get predictions as quickly/cheaply/bestly as possible. TBD what best actually means.
Mesmer segmentation (whole cell segmentation)
DeepCell Pretrained Models Intro: github permalink
Important note on input data:
Data format: The Mesmer model expects two channels of imaging data. The first channel must be a nuclear channel (such as DAPI). The second channel must be a membrane or cytoplasmic channel (such as E-Cadherin).
Notebook: github permalink
Sample data
Angelo Lab ARK
https://github.com/angelolab/ark-analysis
Notebook to run sample data: github permalink
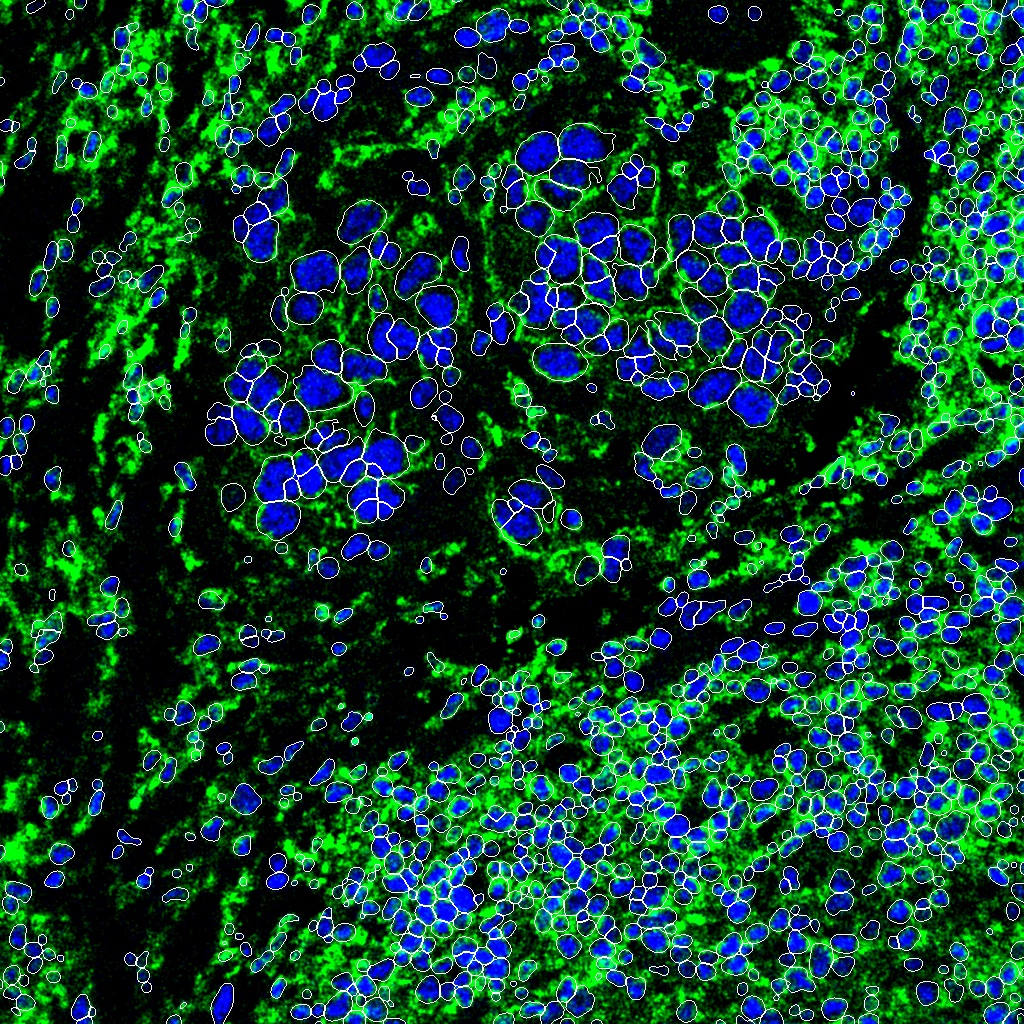
The below images were generated with that notebook run locally over this sample data: HuggingFace. The data includes lots of 1024x1024 cell images.
The notebook preprocesses input channels into an image file that’s sent to DeepCell.
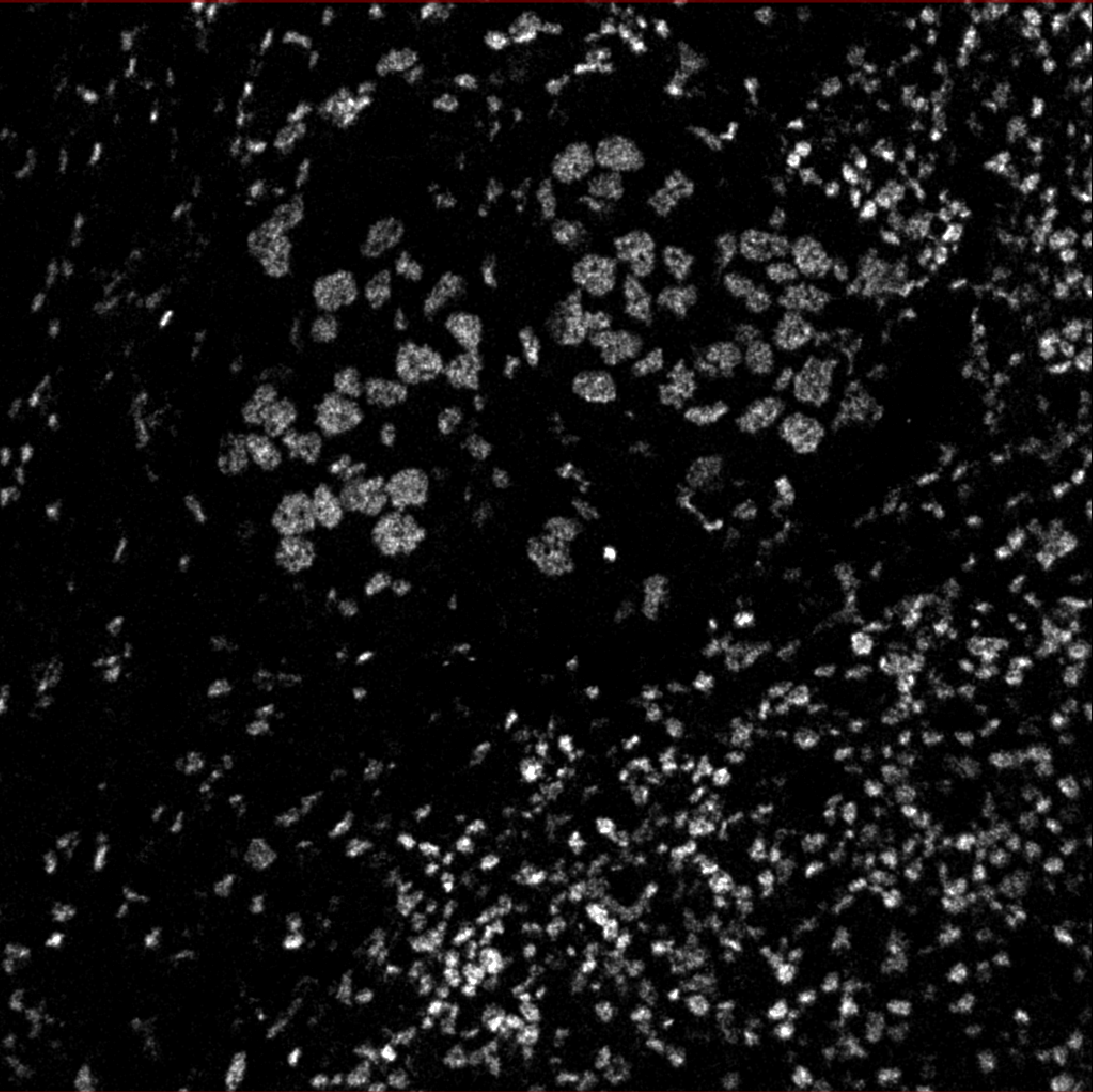
Then DeepCell predicts cell segmentation and sends data back, processed into an image overlay.
10xgenomics
Follow up on viability / usability of this data for Mesmer.
Examples with potential?
Human Prostate Cancer, Adjacent Normal Section with IF Staining (FFPE)
Downloaded tif: Visium_FFPE_Human_Prostate_IF_image.tif, 724.9mb, 26624 x 25088
418mb file?